


 Next: Navsetup
Up: Implementational details
Previous: Implementational details
Contents
Next: Navsetup
Up: Implementational details
Previous: Implementational details
Contents
The physical parameters, information about the grid, precalculated discretizations and some other
stuff is kept in the class
Scene. The most important variables are listened below.
| variable |
description |
| gridp[3] |
This defines the number of cells in every direction for the whole domain 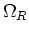 ,
the inner cells are numbered from 1 to gridp[ ,
the inner cells are numbered from 1 to gridp[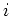 ], ],
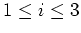 .
The cells which are computed by the actual process are defined in the class
ParParams, which is described in ( .
The cells which are computed by the actual process are defined in the class
ParParams, which is described in (![[*]](file:/usr/share/latex2html/icons/crossref.png) ). ). |
| d[3][] |
This variable contains the spaces between the grid lines in every direction (see
figure ![[*]](file:/usr/share/latex2html/icons/crossref.png) ). For convenience: access is possible using the macros DX[ ). For convenience: access is possible using the macros DX[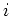 ], DY[ ], DY[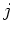 ] and DZ[ ] and DZ[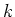 ]. ]. |
| dm[3][] |
This variable contains the spaces between the middles of the cells in every direction
(marked by the circles in figure ![[*]](file:/usr/share/latex2html/icons/crossref.png) ). For convenience:
access is possible using the macros DXM[ ). For convenience:
access is possible using the macros DXM[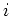 ], DYM[ ], DYM[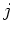 ] and DZM[ ] and DZM[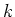 ]. ]. |
| kabs[3][] |
This variable contains the absolute positions of the grid lines. |
| ddstar[3][3][], ddPstar[3][3][], ddSstar[3][3][] |
These variables contain different discretizations for the second derivative, where
the first index selects the direction and the second index selects the appropriate weight (left - 0, middle - 1, right - 2).
The discretizations are described in (![[*]](file:/usr/share/latex2html/icons/crossref.png) ). ). |
| periodbound |
This is a bit field indicating if periodic boundary conditions are set in the
appropriate direction (bit 0 - 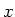 -direction, bit 1 - -direction, bit 1 - 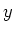 -direction, bit 2 - -direction, bit 2 - 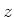 -direction). -direction). |
The 3D matrices, for example velocity and pressure data, are kept in the class
Navier. There are two derived classes,
NavierSetup and
NavierCalc.
The dimensions of these matrices are defined in the following table.
| matrix |
dimension |
flag, P, F, G, H, RHS, T, CH[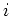 ] ] |
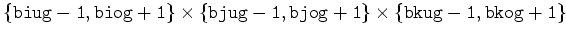 |
| U |
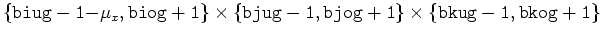 |
| V |
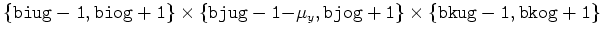 |
| W |
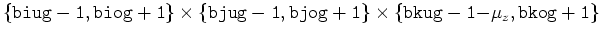 |
The variable
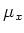
is set to 1 if periodic boundary conditions are set in
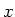
-direction or if the current process
has a neighbour in
south-direction and 0 otherwise. The variables
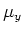
and
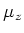
are set in a similar way.
The cells where boundary values need to be set (for one specific parallel process)
are stored in lists which are defined in NavierCalc.
| list |
contains |
| InflowList |
cells where Inflow b.c. are set (parameters are stored in list) |
| SlipList |
cells where Slip b.c. (Neumann) are set |
| NoslipList |
cells where Noslip b.c. (Dirichlet) are set |
| PdpdnList[color][face] |
cells where prescribed boundary conditions (b.c.) lead to homogeneous
Neumann b.c. for the pressure and which have the color color. These cells have exactly
one fluid cell neighbouring at face face. |
| PSpecdpdnList[color] |
cells where prescribed boundary conditions (b.c.) lead to homogeneous
Neumann b.c. for the pressure and which have the color color. These cells have more
than one neighbouring fluid cell. This requires some more involved boundary treatment. |
| PInOut2List[color][face] |
cells with the prescribed outflow condition 2,
it is assumed that no cells with more than one neighbouring fluid
cell exist.
Some special boundary treatment is required. |
| PInOut1List[color][face] |
same as PInOut2List for outflow condition 1. |



 Next: Navsetup
Up: Implementational details
Previous: Implementational details
Contents
Martin Engel
2004-03-15
Next: Navsetup
Up: Implementational details
Previous: Implementational details
Contents
Martin Engel
2004-03-15