


 Next: Implementational details
Up: Navcalc and Navcalcmpi
Previous: Command line arguments
Contents
Next: Implementational details
Up: Navcalc and Navcalcmpi
Previous: Command line arguments
Contents
Parallelization issues
For the parallelization the domain
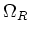
is divided into several rectangular subdomains
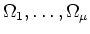
.
A simple 2D example is shown in figure
![[*]](file:/usr/share/latex2html/icons/crossref.png)
.
Figure:
2D parallelization example.
|
|
Each process holds some ghost cells which overlap inner cells of the adjacent process.
Values are copied from these to the ghost cells when necessary.
To minimize communication, the program divides
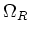
in a way that minimizes
the area of the touching faces and equilibrates the number of cells in the different subdomains.
The data required for parallelization is kept in the class ParParams
which can be found in parallel.hpp. The most important variables are described in the following table:
| variable |
description |
| biug, biog, bjug, bjog, bkug, bkog |
This defines the computational domain for this process
(excluding the necessary ghost cells) as
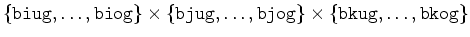 .
For your convenience, read access is possible using the macros iug, iog, etc.
For example, .
For your convenience, read access is possible using the macros iug, iog, etc.
For example,
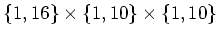 will be divided
into will be divided
into
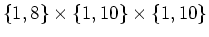 and and
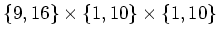 . . |
| me |
Process ID of this task. |
| tn, ts, tw, te, tb, tt |
IDs of the neighbour processes in each direction (see figure ![[*]](file:/usr/share/latex2html/icons/crossref.png) ).
The value -1 means that there is no neighbour process. ).
The value -1 means that there is no neighbour process. |



 Next: Implementational details
Up: Navcalc and Navcalcmpi
Previous: Command line arguments
Contents
Martin Engel
2004-03-15
Next: Implementational details
Up: Navcalc and Navcalcmpi
Previous: Command line arguments
Contents
Martin Engel
2004-03-15
![\includegraphics[width=0.5\textwidth,keepaspectratio]{p4.eps}](img136.png)